Online Quantitative Transcriptome Analysis
The current revolution in sequencing technologies allows us to obtain a much more detailed picture of transcriptomes. Studying them under understanding of the underlying mechanisms of gene expression and processing. An important prerequisite is to be able to accurately determine the full complement of RNA transcripts and to infer their abundance various limitations and biases in next-generation sequencing (NGS) technologies.
We present a Machine Learning powered platform for quantitatively analyzing RNA-seq experiments, called oqtans. Oqtans is integrated in the easy-to-use Galaxy framework [1] and builds on NGS analysis methods developed in the Rätsch Laboratory and by others, which can be freely combined.
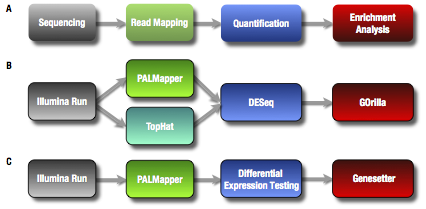
[A] General RNAseq workflow. [B, C] Example workflows using different tools.
Oqtans also allows for tools to be compared against each other.
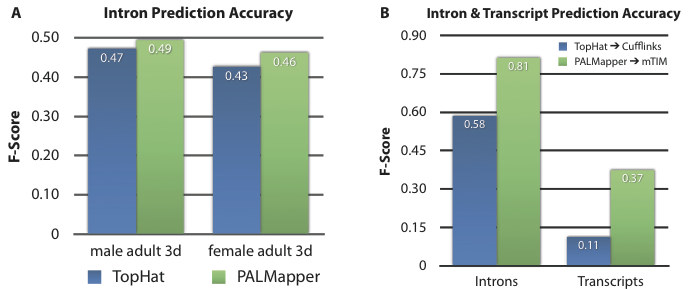
Some of our own tools integrated into oqtans:
- PALMapper [2] is a short read mapper which efficiently computes both unspliced and spliced alignments at high accuracy by taking advantage of base quality information and computational splice site predictions.
- mTiM [3] is a machine learning-based transcript reconstruction method, which exploits features derived from RNA-seq read alignments and from computational splice sites predictions to infer the exon-intron structure of the corresponding transcripts.
- rQuant [4] is a method based on quadratic programming that simultaneously estimates biases inherent in library preparation, sequencing, and read mapping and accurately determines the abundances of given transcripts.
- rDiff [5] [6] is a set of statistical test techniques that determine significant differences between two RNA-Seq experiments to find differentially expressed regions with or without knowledge of transcripts.
An overview over all tools can be found on the Oqtans Tools Page. The machine-learning tools above show very accurate results and perform better or on par with the state-of-the-art for short read alignments, transcript identification and quantification as well as differential expression analysis. Their combination into a powerful workflow integrated in the Galaxy framework makes it possible to easily and effectively conduct a complete quantitative RNA-Seq analysis.
It can be accessed on our server, locally or in the cloud:
http://galaxy.cbio.mskcc.org (public compute server)
http://cloud.oqtans.org (online demo instance)
cm-ba5c56b95144e564f70e5762dc5fa177/shared/2013-11-07--22-16/ (share-an-instance-cluster for Galaxy CloudMan)
Our programs are free, open source and standalone, and can be downloaded from their respective Supplement page (links to the left).
Oqtans Installation
You can find installation instructions at the Oqtans Installation Page and instructions how to get started using the Amazon cloud image (ami-65376a0c) can be found on the Oqtans Instantiation Page.
Oqtans Use Cases
In the oqtans publication, we demonstrate two RNAseq analysis workflows using a variety of tools. These are compiled on the Oqtans Use Cases page.
Materials
- A Multifunctional Workbench for RNA-seq Data Analysis poster presented at High Throughput Sequencing HiTSeq 2013 conference July 19-20, 2013, Berlin, Germany.
- The slides of the talk Reproducible Quantitative Transcriptome Analysis with oqtans presented at Bioinformatics Open Source Conference(BOSC2013) July 19-20, 2013, Berlin, Germany.
- A poster about Online Transcriptome Analysis with Galaxy presented at Computational Biology Annual Retreat, Mohonk Mountain house, New York September 17-18, 2012.
- The slides of the talk Easier Workflows & Tool Comparison with Oqtans+ by Vipin T Sreedharan at Galaxy Community Conference July 26, 2012, University of Illinois, Chicago.
- The slides of Part I and Part II of a workshop presented at Meeting on Advances and Challenges of RNA-Seq Analysis in Halle, Germany in June 2012.
- The abstract presenting Oqtans at the Technology Track of ISMB/ECCB 2011.
- The handout distributed at the Technology Track of ISMB/ECCB 2011 and presenting oqtans.
- The slides of the presentation at the Technology Track of ISMB/ECCB 2011.
- The slides. of the presentation by Sebastian J Schultheiss at the 7th Student Council Symposium 2011 (Winner of the 2nd place Best Oral Presentation Award).
- A poster presented at ISMB 2010 describing Oqtans.
- The slides of the presentation Online Quantitative Transcriptome Analysis by Gunnar Rätsch at Hitseq 2010 in Boston.
Contact
Please email us at support@oqtans.org
Developers contributing to Oqtans
Regina Bohnert, Philipp Drewe, Nico Goernitz, Géraldine Jean, André Kahles, Pramod Mudrakarta, Gunnar Rätsch, Sebastian J. Schultheiss, Vipin T. Sreedharan, Georg Zeller
References
| [1] | Blankenberg, D. et al., Galaxy: a web-based genome analysis tool for experimentalists, Curr Protoc Mol Biol, 2010. |
| [2] | Jean, G. et al., RNA-seq read alignments with PALMapper, Curr Protoc Bioinform, 2010. |
| [3] | Zeller, G. et al., mTiM: Rapid and accurate transcript reconstruction from RNA-seq data. Technical Report arXiv:1309.5211, arXiv, 2013. |
| [4] | Bohnert, R. et al., rQuant.web: a tool for RNA-Seq-based transcript quantitation, NAR, 2010. |
| [5] | Stegle, O. et al., Statistical tests for detecting differential RNA-transcript expression from read counts. Nature Precedings, 2010. |
| [6] | Drewe, P. et al., rDiff : Accurate Detection of Differential RNA Processing. Nucl. Acids Res. (2013) 41 (10): 5189-5198. |