Members from our lab are part of an interdisciplinary research team of biomedical and computer scientists from ETH Zurich, the
University of Zurich, and the University Hospital Zurich. They have now developed a machine learning approach that allows such cell changes and drug effects to be modelled and predicted with much greater accuracy and nuance than before.
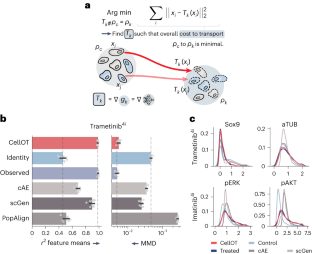
Abstract Understanding and predicting molecular responses in single cells upon chemical, genetic or mechanical perturbations is a core question in biology. Obtaining single-cell measurements typically requires the cells to be destroyed. This makes learning heterogeneous perturbation responses challenging as we only observe unpaired distributions of perturbed or non-perturbed cells. Here we leverage the theory of optimal transport and the recent advent of input convex neural architectures to present CellOT, a framework for learning the response of individual cells to a given perturbation by mapping these unpaired distributions. CellOT outperforms current methods at predicting single-cell drug responses, as profiled by scRNA-seq and a multiplexed protein-imaging technology. Further, we illustrate that CellOT generalizes well on unseen settings by (1) predicting the scRNA-seq responses of holdout patients with lupus exposed to interferon-β and patients with glioblastoma to panobinostat; (2) inferring lipopolysaccharide responses across different species; and (3) modeling the hematopoietic developmental trajectories of different subpopulations.
Resources
Nature Methods Paper
Nature Methods Research Briefing
Press
ETH News
Watson
idw
Blick
nau.ch
punkt4