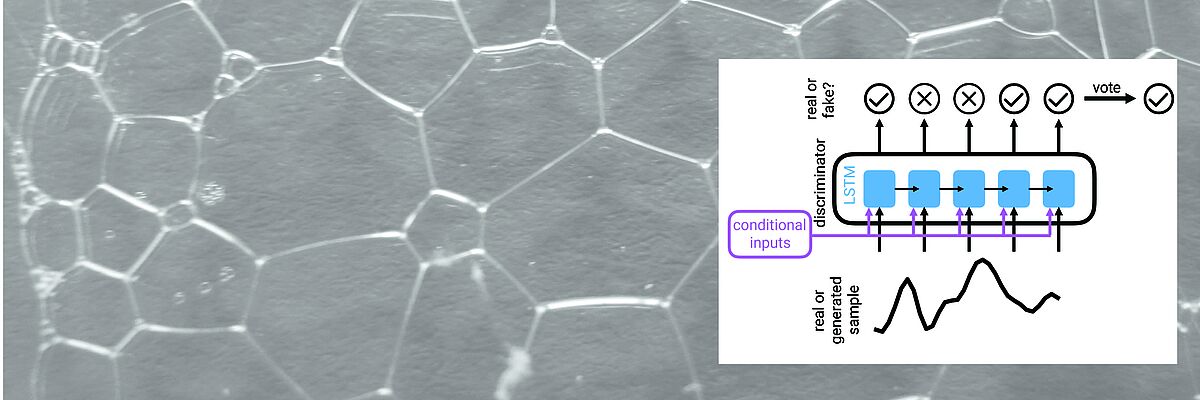
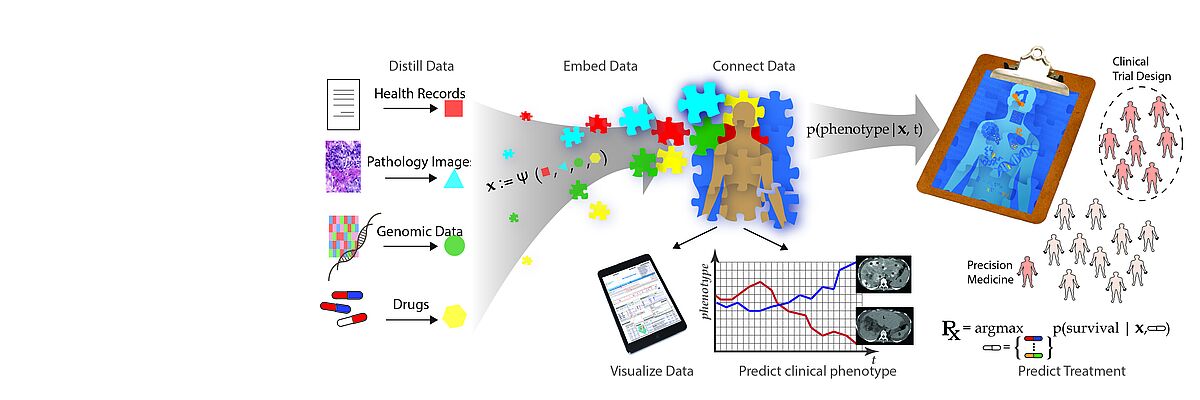
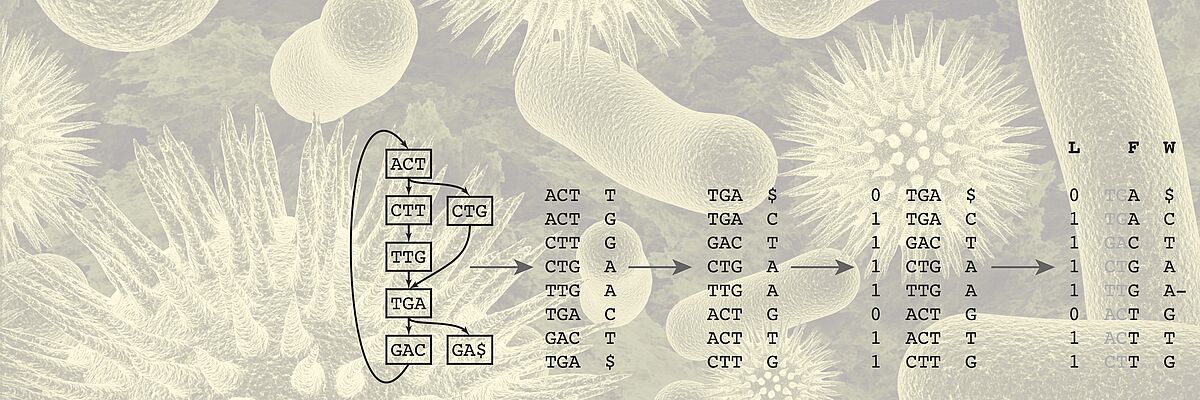
The BMI lab bridges research in Machine Learning and Sequence Analysis methodology research and its application to biomedical problems. We collaborate with biologists and clinicians to develop real-world solutions.
We work on research questions and foundational challenges in storing, analysing, and searching extensive heterogeneous and temporal data, especially in the biomedical domain. Our lab members address technical and non-technical research questions in collaboration with biologists and clinicians. At the research group’s core is an active knowledge exchange in both directions between the methods and the application-driven researchers.
The emergence of data-driven medicine leverages data and algorithms to shape how we diagnose and treat patients. Machine Learning approaches allow us to capitalise on the vast amount of data produced in clinical settings to generate novel biomedical insights and build more precise predictive models of disease outcomes and treatment efficacy.
We work towards this transformation mainly but not exclusively in two key areas. One key application area is the analysis of heterogeneous data of cancer patients. For Genomics, we develop algorithms for storing, compressing, and searching extensive genomics datasets. Another key area is the development of time series models of patient health states and early warning systems for intensive care units.
Publications
Abstract Clinical time series data are critical for patient monitoring and predictive modeling. These time series are typically multivariate and often comprise hundreds of heterogeneous features from different data sources. The grouping of features based on similarity and relevance to the prediction task has been shown to enhance the performance of deep learning architectures. However, defining these groups a priori using only semantic knowledge is challenging, even for domain experts. To address this, we propose a novel method that learns feature groups by clustering weights of feature-wise embedding layers. This approach seamlessly integrates into standard supervised training and discovers the groups that directly improve downstream performance on clinically relevant tasks. We demonstrate that our method outperforms static clustering approaches on synthetic data and achieves performance comparable to expert-defined groups on real-world medical data. Moreover, the learned feature groups are clinically interpretable, enabling data-driven discovery of task-relevant relationships between variables.
Authors Fedor Sergeev, Manuel Burger, Polina Leshetkina, Vincent Fortuin, Gunnar Rätsch, Rita Kuznetsova
Submitted ML4H 2025 (PMLR)
Abstract CRISPR-based genetic perturbation screens paired with single-cell transcriptomic readouts (Perturb-seq) offer a powerful tool for interrogating biological systems. Yet the resulting datasets are heterogeneous—particularly in vivo—and currently used cell-level perturbation labels reflect only CRISPR guide RNA exposure rather than perturbation state; further, many perturbations have a minimal effect on gene expression. For perturbations that do alter the transcriptomic state of cells, intracellular guide RNA abundance exhibits a dose-response association with perturbation efficacy. We combine (i) per-perturbation, expression-only classifiers trained with non-negative negative–unlabeled (nnNU) risk to yield calibrated scores reflecting the perturbation state of single cells and (ii) a monotone guide abundance prior to yield a per-cell pseudo-posterior that supports both assignment of perturbation probability and selection of affected gene features. To obtain a low-dimensional representation that allows for the accurate reconstruction of gene-level marginals for counterfactual decoding, we train an autoencoder with a quantile–hurdle reconstruction loss and feature-weighted emphasis on perturbation-affected genes. The result is a perturbation-aware latent embedding amenable to downstream trajectory modeling (e.g., optimal transport or flow matching) and a principled probability of perturbation for each non-control cell derived jointly from its guide counts and transcriptome.
Authors Florian Hugi, Tanmay Tanna, Randall J. Platt, Gunnar Rätsch
Submitted NeurIPS 2025 AI4D3
Abstract Spot-based spatial transcriptomics (ST) technologies like 10x Visium quantify genome-wide gene expression and preserve spatial tissue organization. However, their coarse spot-level resolution aggregates signals from multiple cells, preventing accurate single-cell analysis and detailed cellular characterization. Here, we present DeepSpot2Cell, a novel DeepSet neural network that leverages pretrained pathology foundation models and spatial multi-level context to effectively predict virtual single-cell gene expression from histopathological images using spot-level supervision. DeepSpot2Cell substantially improves gene expression correlations on a newly curated benchmark we specifically designed for single-cell ST deconvolution and prediction from H&E images. The benchmark includes 20 lung, 7 breast, and 2 pancreatic cancer samples, across which DeepSpot2Cell outperformed previous super-resolution methods, achieving respective improvements of 46%, 65%, and 38% in cell expression correlation for the top 100 genes. We hope that DeepSpot2Cell and this benchmark will stimulate further advancements in virtual single-cell ST, enabling more precise delineation of cell-type-specific expression patterns and facilitating enhanced downstream analyses. Code availability: https://github.com/ratschlab/DeepSpot
Authors Kalin Nonchev, Glib Manaiev, Viktor H Koelzer, Gunnar Rätsch
Submitted NeurIPS 2025 Imageomics
Abstract Histopathology refers to the microscopic examination of diseased tissues and routinely guides treatment decisions for cancer and other diseases. Currently, this analysis focuses on morphological features but rarely considers gene expression information, which can add an important molecular dimension. Here, we introduce SpotWhisperer, an AI method that links histopathological images to spatial gene expression profiles and their text annotations, enabling molecularly grounded histopathology analysis through natural language. Our method outperforms pathology vision-language models on a newly curated benchmark dataset, dedicated to spatially resolved H&E annotation. Integrated into a web interface, SpotWhisperer enables interactive exploration of cell types and disease mechanisms using free-text queries with access to inferred spatial gene expression profiles. In summary, SpotWhisperer analyzes cost-effective pathology images with spatial gene expression and natural-language AI, demonstrating a path for routine integration of microscopic molecular information into histopathology.
Authors Moritz Schaefer, Kalin Nonchev, Animesh Awasthi, Jake Burton, Viktor H Koelzer, Gunnar Rätsch, Christoph Bock
Submitted ICML 2025 FM4LS
Abstract Spatial transcriptomics technology remains resource-intensive and unlikely to be routinely adopted for patient care soon. This hinders the development of novel precision medicine solutions and, more importantly, limits the translation of research findings to patient treatment. Here, we present DeepSpot, a deep-set neural network that leverages recent foundation models in pathology and spatial multi-level tissue context to effectively predict spatial transcriptomics from H&E images. DeepSpot substantially improved gene correlations across multiple datasets from patients with metastatic melanoma, kidney, lung, or colon cancers as compared to previous state-of-the-art. Using DeepSpot, we generated 1 792 TCGA spatial transcriptomics samples (37 million spots) of the melanoma and renal cell cancer cohorts. We anticipate this to be a valuable resource for biological discovery and a benchmark for evaluating spatial transcriptomics models. We hope that DeepSpot and this dataset will stimulate further advancements in computational spatial transcriptomics analysis.
Authors Kalin Nonchev, Sebastian Dawo, Karina Selina, Holger Moch, Sonali Andani, Tumor Profiler Consortium, Viktor Hendrik Koelzer, Gunnar Rätsch
Submitted MedRxiv